Excel autocorrect errors still plague genetic research, raising concerns over scientific rigour
- Written by Mark Ziemann, Lecturer in Biotechnology and Bioinformatics, Deakin University
Autocorrection, or predictive text, is a common feature of many modern tech tools, from internet searches to messaging apps and word processors. Autocorrection can be a blessing, but when the algorithm makes mistakes it can change the message in dramatic and sometimes hilarious ways.
Our research shows autocorrect errors, particularly in Excel spreadsheets, can also make a mess of gene names in genetic research. We surveyed more than 10,000 papers with Excel gene lists published between 2014 and 2020 and found more than 30%[1] contained at least one gene name mangled by autocorrect.
This research follows our 2016 study that found around 20%[2] of papers contained these errors, so the problem may be getting worse. We believe the lesson for researchers is clear: it’s past time to stop using Excel and learn to use more powerful software.
Excel makes incorrect assumptions
Spreadsheets apply predictive text to guess what type of data the user wants. If you type in a phone number starting with zero, it will recognise it as a numeric value and remove the leading zero. If you type “=8/2”, the result will appear as “4”, but if you type “8/2” it will be recognised as a date.
With scientific data, the simple act of opening a file in Excel with the default settings can corrupt the data due to autocorrection. It’s possible to avoid unwanted autocorrection if cells are pre-formatted prior to pasting or importing data, but this and other data hygiene tips aren’t widely practised.
In genetics, it was recognised way back in 2004[3] that Excel was likely to convert about 30 human gene and protein names to dates. These names were things like MARCH1, SEPT1, Oct-4, jun, and so on.
Several years ago, we spotted this error in supplementary data files attached to a high impact journal article and became interested in how widespread these errors are. Our 2016 article indicated that the problem affected middle and high ranking journals at roughly equal rates. This suggested to us that researchers and journals were largely unaware of the autocorrect problem and how to avoid it.
As a result of our 2016 report, the Human Gene Name Consortium, the official body responsible for naming human genes, renamed the most problematic genes. MARCH1 and SEPT1 were changed to MARCHF1 and SEPTIN1 respectively, and others had similar changes.
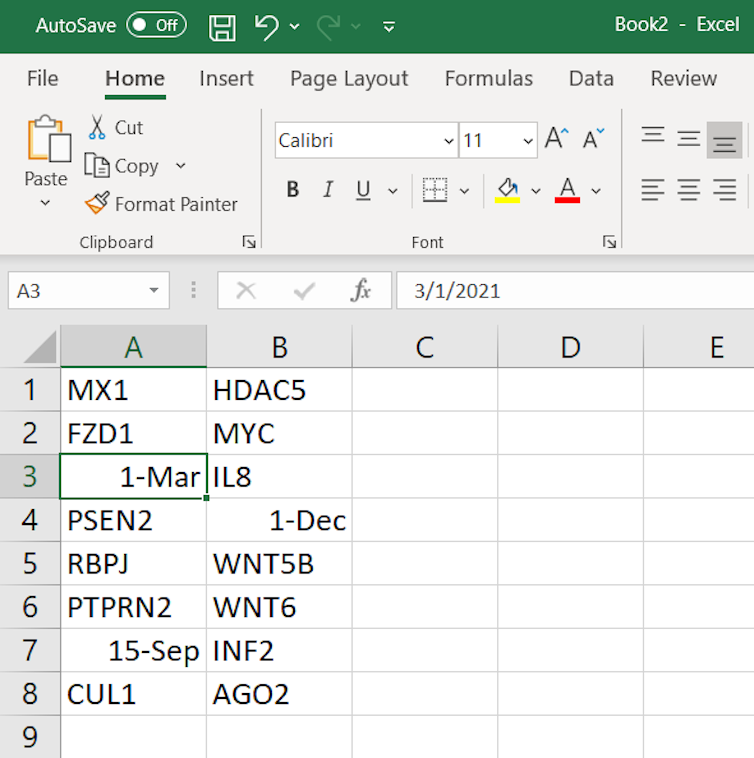 An example list of gene names in Excel.
An example list of gene names in Excel.
An ongoing problem
Earlier this year we repeated our analysis. This time we expanded it to cover a wider selection of open access journals, anticipating researchers and journals would be taking steps to prevent such errors appearing in their supplementary data files.
We were shocked to find in the period 2014 to 2020 that 3,436 articles, around 31% of our sample, contained gene name errors[4]. It seems the problem has not gone away, and is actually getting worse.
Small errors matter
Some argue these errors don’t really matter, because 30 or so genes is only a small fraction of the roughly 44,000 in the entire human genome, and the errors are unlikely to overturn to conclusions of any particular genomic study.
Anyone reusing these supplementary data files will find this small set of genes missing or corrupted. This might be irritating if your research project examines the SEPT gene family, but it’s just one of many gene families in existence.
We believe the errors matter because they raise questions about how these errors can sneak into scientific publications. If gene name autocorrect errors can pass peer-review undetected into published data files, what other errors might also be lurking among the thousands of data points?
Spreadsheet catastrophes
In business and finance, there are many examples where spreadsheet errors led to costly and embarrassing losses[5].
In 2012, JP Morgan declared a loss of more than US$6 billion thanks to a series of trading blunders made possible by formula errors[6] in its modelling spreadsheets. Analysis of thousands of spreadsheets at Enron Corporation, from before its spectacular downfall in 2001, show almost a quarter contained errors[7].
A now-infamous article by Harvard economists Carmen Reinhart and Kenneth Rogoff was used to justify austerity cuts in the aftermath of the global financial crisis, but the analysis contained a critical Excel error[8] that led to omitting five of the 20 countries in their modelling.
Read more: The Reinhart-Rogoff error – or how not to Excel at economics[9]
Just last year, a spreadsheet error at Public Health England[10] led to the loss of data corresponding to around 15,000 positive COVID-19 cases. This compromised contact tracing efforts for eight days while case numbers were rapidly growing. In the health-care setting, clinical data entry errors[11] into spreadsheets can be as high as 5%, while a separate study of hospital administration spreadsheets[12] showed 11 of 12 contained critical flaws.
In biomedical research, a mistake in preparing a sample sheet resulted in a whole set of sample labels being shifted by one position and completely changing the genomic analysis results[13]. These results were significant because they were being used to justify the drugs patients were to receive in a subsequent clinical trial. This may be an isolated case, but we don’t really know how common such errors are in research because of a lack of systematic error-finding studies.
Better tools are available
Spreadsheets are versatile and useful, but they have their limitations. Businesses have moved away from spreadsheets to specialised accounting software, and nobody in IT would use a spreadsheet to handle data when database systems such as SQL are far more robust and capable.
However, it is still common for scientists to use Excel files to share their supplementary data online. But as science becomes more data-intensive and the limitations of Excel become more apparent, it may be time for researchers to give spreadsheets the boot.
In genomics and other data-heacy sciences, scripted computer languages such as Python and R are clearly superior to spreadsheets. They offer benefits including enhanced analytical techniques, reproducibility, auditability and better management of code versions and contributions from different individuals. They may be harder to learn initially, but the benefits to better science are worth it in the long haul.
Excel is suited to small-scale data entry and lightweight analysis. Microsoft says[14] Excel’s default settings are designed to satisfy the needs of most users, most of the time.
Clearly, genomic science does not represent a common use case. Any data set larger than 100 rows is just not suitable for a spreadsheet.
Researchers in data-intensive fields (particularly in the life sciences) need better computer skills. Initiatives such as Software Carpentry[15] offer workshops to researchers, but universities should also focus more on giving undergraduates the advanced analytical skills they will need.
References
- ^ more than 30% (journals.plos.org)
- ^ around 20% (genomebiology.biomedcentral.com)
- ^ 2004 (bmcbioinformatics.biomedcentral.com)
- ^ gene name errors (journals.plos.org)
- ^ costly and embarrassing losses (www.eusprig.org)
- ^ formula errors (qz.com)
- ^ almost a quarter contained errors (ieeexplore.ieee.org)
- ^ critical Excel error (theconversation.com)
- ^ The Reinhart-Rogoff error – or how not to Excel at economics (theconversation.com)
- ^ spreadsheet error at Public Health England (www.bbc.com)
- ^ clinical data entry errors (bmjopen.bmj.com)
- ^ study of hospital administration spreadsheets (www.igi-global.com)
- ^ completely changing the genomic analysis results (www.nature.com)
- ^ Microsoft says (www.bbc.com)
- ^ Software Carpentry (software-carpentry.org)

















